You are here
June 30, 2014
Molecular Insights into a Target for Multidrug Resistance
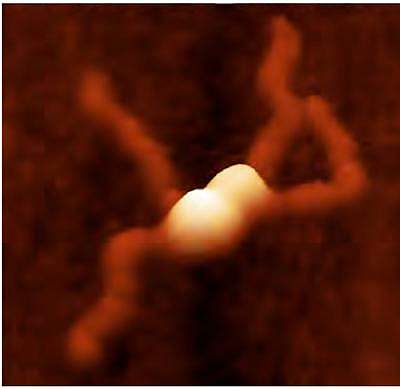
Scientists deciphered the structure and mechanism of a protein that bacteria need to spread the genes for antibiotic resistance. The findings could enable the development of novel drugs to combat multidrug-resistant bacteria.
Antibiotics disrupt bacteria’s ability to grow and replicate. Microbes that are resistant to antibiotics are a growing public health problem. Some bacteria, including strains of Staphylococcus aureus (Staph), have become extremely difficult to treat because the microbes are able to survive different antibiotics. Decades of widespread use have encouraged the spread of bacteria with resistance to multiple antibiotics.
Genes that confer drug resistance can be passed between bacteria on small circular DNA molecules called plasmids. Plasmids copy themselves separately from the bacterial chromosome by using the host bacterium’s DNA replication machinery. Past studies have found that most plasmids carrying drug-resistant genes in S. aureus encode a highly conserved protein called RepA that’s part of the complex needed to begin plasmid replication.
A team at Duke University Medical Center led by Dr. Maria Schumacher and colleagues at the University of Sydney in Australia investigated the structure and mechanism of RepA. Their study was funded in part by NIH’s National Institute of General Medical Sciences (NIGMS) and National Center for Research Resources (NCRR). Results appeared online on June 9, 2014, in Proceedings of the National Academy of Sciences.
RepA has 3 main domains: a highly conserved N-terminal domain (NTD) that binds DNA, a long and variable linker region, and a C-terminal domain (CTD) that interacts with other proteins involved in replication. The scientists used a technique called X-ray crystallography to determine the structure of the NTD and CTD. They found that NTDs were configured to form dimers and tetramers with themselves.
Next, the scientists studied how the NTD associates with DNA. They determined that RepA binds along interon boxes—sites where plasmid replication begins—next to each other. This side-by-side configuration is associated with DNA melting, the opening up of the double-stranded DNA molecule and one of the initial steps of replication.
Modeling suggested that 2 separate DNA strands are linked together by NTD tetramers. To take a closer look at the molecular level, the scientists used a technique called atomic force microscopy. The results confirmed that NTD tetramers stick together to “handcuff” 2 DNA plasmids together.
Plasmids left to replicate uncontrolled can harm host cells by overburdening the cells’ metabolism. The handcuffing structure suggests a molecular mechanism for controlling how many plasmid copies can be made in the bacterium; the bridging structure can hinder plasmid replication by preventing the initiation of replication.
“If plasmids can’t replicate, they go away,” Schumacher says. These results give new molecular insight into how to design drugs to target RepA and prevent multidrug resistant Staph from spreading resistance genes.
—by Shu Hui Chen, Ph.D. and Harrison Wein, Ph.D.
Related Links
- Method Quickly Assesses Antibiotics
- New Drug Effective Against MRSA in Mice
- Antimicrobial (Drug) Resistance
- Methicillin-Resistant Staphylococcus aureus (MRSA)
References: Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein. Schumacher MA,Tonthat NK,Kwong SM,Chinnam NB,Liu MA,Skurray RA,Firth N. Proc Natl Acad Sci U S A.2014 Jun 9. pii: 201406065. [Epub ahead of print]. PMID: 24927575.
Funding: NIH’s National Institute of General Medical Sciences (NIGMS) and National Center for Research Resources (NCRR); the U.S. Department of Energy, and the National Health and Medical Research Council of Australia.
