You are here
March 23, 2015
Novel Approach Gives Insights Into Tumor Development
At a Glance
- Scientists used a powerful new technique to turn off all the genes in mouse lung cancer cells and test how they affect tumor growth and metastasis.
- The study provides a roadmap for understanding how tumors evolve over time.
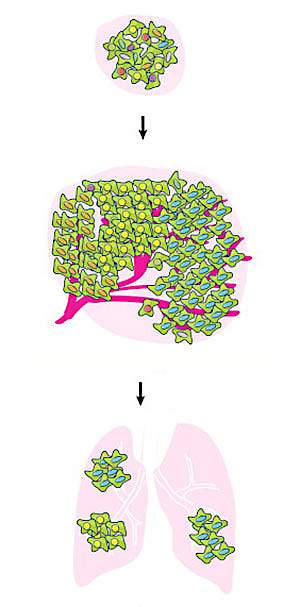
Lung cancer is the most common cause of cancer deaths both nationwide and worldwide. Past studies have found many genetic alterations involved in lung cancer development. However, tumor growth and metastasis occurs amid complex, evolving, and diverse genetic changes. A major challenge in understanding the cancer genome is to identify the genetic changes that specifically drive the process of tumor development.
To systematically test the effects of mutations across the cancer genome, researchers at the Massachusetts Institute of Technology and their colleagues used a technique called a CRISPR-Cas screen to disrupt specific sequences. Bacteria use the CRISPR-Cas system to protect themselves from invaders. Short stretches of genetic material from bacteriophages (viruses that attack bacteria) and plasmids (pieces of DNA exchanged between bacteria) are incorporated into the system to detect targets for destruction in the future.
For target sequences, the scientists used a pooled genome-wide mouse library of more than 67,000 short genetic sequences called single-guide RNAs, or sgRNAs. The library included more than 20,000 protein-coding genes and almost 1,200 microRNA sequences, which help regulate gene expression and have been implicated in various cancers. Using a cell line derived from a mouse non-small-cell lung cancer, they added the CRISPR-Cas system genes and then the sgRNA library. The work, led by Drs. Feng Zhang and Phillip Sharp, was funded by NIH’s National Cancer Institute (NCI) and others. Results were published on March 12, 2015, in Cell.
When injected under the skin of immunocompromised mice, the collection of mutant cells formed tumors at the injection site that metastasized to the lungs. Late primary tumors contained only a fraction of all the sgRNAs in the collection—less than 4%. Metastases were dominated by a very small number of sgRNAs—on average, only 3.4 per lung. These results suggested that cells with greater ability to proliferate dominated the tumors over time, and that metastases were seeded by a small set of cells.
The team tested a more select pool of sgRNAs from lung metastases in the genome-wide screen. This approach allowed them to quickly test multiple sgRNAs and see how the mutations competed. Mice transplanted with this subpool had a dramatically higher rate of lung metastasis. The metastases retained only a small fraction of sgRNAs, suggesting that these sgRNAs can outcompete others during tumor growth and metastasis.
Mutations in many of the identified genes have been implicated in human cancers. When the team analyzed gene expression in metastatic tumors from patients with non-small-cell lung cancer, most of the genes (up to 75%) were downregulated. Future sequencing of patient metastases may further connect the genes identified in this study to the clinic.
“Tumor evolution is an extremely complex set of processes, or hallmarks, controlled by networks of genes,” Sharp says. Identifying the changes that are important for tumor development could lead to more targeted therapies, better prevention, and new detection strategies.
—by Harrison Wein, Ph.D.
Related Links
- Method Can Target Specific Microbes
- Gene Changes Identified in Most Common Lung Cancer
- Lung Cancer
- The Cancer Genome Atlas
References: Genome-wide CRISPR Screen in a Mouse Model of Tumor Growth and Metastasis. Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, Lee H, Zhang F, Sharp PA. Cell. 2015 Mar 12;160(6):1246-1260. doi: 10.1016/j.cell.2015.02.038. Epub 2015 Mar 5. PMID: 25748654.
Funding: NIH’s National Cancer Institute (NCI), National Institute of Mental Health (NIMH), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), and National Human Genome Research Institute (NHGRI); Marie D. and Pierre Casimir-Lambert Fund; Skolkovo Foundation; National Science Foundation; Keck Foundation; New York Stem Cell Foundation; Damon Runyon Foundation; Searle Scholars Foundation; Merkin Foundation; Vallee Foundation; Dale Frey Award for Breakthrough Scientists; Simons Center for the Social Brain; and U.S. Department of Defense.
