You are here
September 16, 2013
Technique Visualizes Brain Tumors During Surgery
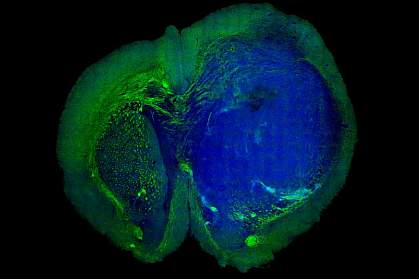
A new method can distinguish tumors from normal brain tissue in living mice. With further refinement, the approach could help doctors remove brain tumors with great precision.
Recognizing the difference between tumors and normal brain tissue during surgery is a major challenge. Removing healthy tissue can cause neurologic problems, but leaving tumor tissue behind can allow the cancer to spread again. This is a particular problem with glioblastoma multiforme, the most common form of malignant brain cancer in adults. Glioblastoma tumors grow quickly and are difficult to treat. The tumors infiltrate normal brain tissue and can’t be easily singled out.
Experimental methods to tell the difference between tumors and normal tissue during surgery have had limited success. Over the past 15 years, a team led by Dr. Sunney Xie at Harvard University has been developing a technique called stimulated Raman scattering (SRS) microscopy. The method takes advantage of the fact that chemical bonds in molecules have their own sets of vibration frequencies, and produce unique patterns of scattered light called Raman spectra. These spectra can be used as fingerprints to identify and differentiate different molecules in a complex environment. SRS microscopy involves shining noninvasive lasers to excite particular Raman frequencies in tissues. The weak light signals emitted by the tissues vary depending on the tissues’ molecular composition, such as lipids, proteins, and DNA.
In collaboration with Dr. Daniel Orringer and colleagues at the University of Michigan Medical School, Xie’s team applied SRS microscopy to the problem of distinguishing protein-rich glioblastomas from more lipid-rich surrounding tissue. Their work was funded by an NIH Director’s Transformative Research Award and by NIH’s National Cancer Institute (NCI). The results appeared on September 4, 2013, in Science Translational Medicine.
By combining SRS images made from light at 2 different frequencies, the scientists were able to construct images that identified tissues with different lipid and protein content. To test the approach on tumors, they implanted human glioblastoma cells into mice and allowed them to grow into tumors. They then placed samples on slides and used SRS microscopy to make 2-color images of the samples. For comparison, they froze the samples and stained them with hematoxylin and eosin (H&E), the current approach used to diagnose brain tumors.
The scientists found that SRS microscopy worked as well as H&E in distinguishing tumor-infiltrated brain tissue from surrounding healthy tissue. They then adapted the technique for use in live mice. Craniectomies exposed the tumor and adjacent brain tissue for SRS imaging. While standard microscopy found no obvious evidence of the tumor, SRS microscopy identified regions with extensive tumor infiltration.
“For more than 100 years, hematoxylin and eosin stain has been the gold standard for this type of imaging,” Xie says. “But with this [SRS] technology, we don't need to freeze the tissue, we don't need to stain tissue, and we don't need to biopsy — this acts like an optical biopsy and allows us to identify the tumor margins at a cellular level.”
Several challenges remain before SRS microscopy could be used in the clinic. These include the engineering challenge of making a handheld surgical device with motion correction to acquire images from within a surgical cavity.
— by Harrison Wein, Ph.D.
Related Links
References: Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci Transl Med. 2013 Sep 4;5(201):201ra119. doi: 10.1126/scitranslmed.3005954. PMID: 24005159.
Funding: NIH Director’s Transformative Research Award Program and National Cancer Institute (NCI).
